Efficient Pandas: ways to save space and time
The first step to reduce memory usage is to load only columns that are necessary for analysis by utilizing usecols argument in pd.read_csv(): pd.read_csv(url, usecols=[column names/indexes]).
The second – is to specify column types.
When Pandas read a .csv or .json file, the column types are inferred and are set to the largest data type (int64, float64, object). This results in high memory usage and is highly inefficient for big datasets. So, using the right data types can reduce memory usage for Pandas DataFrame
Let’s see how using correct datatypes can improve performance.
For this, we are going to use the FrogID dataset 2.0. FrogID is an Australian national citizen science project that is helping researchers to better understand species diversity, distribution and breeding habitats of frogs. You can join the project with the Android or Apple app. If you are interested in the scientific outcomes of the data analysis, they can be found on the science page of the FrogID project.
import pandas as pd
url = 'https://d2pifd398unoeq.cloudfront.net/FrogIDYear1and2_final_dataset.csv'
cols = ['scientificName', 'sex', 'lifestage', 'eventDate', 'stateProvince', 'geoprivacy', 'recordedBy']
df = pd.read_csv(frogs_fn, usecols=cols)
Object dtype
Object dtype is often a great candidate for conversion to category data. Object is roughly comparable to str in native Python. As strings occupy a significant amount of space, converting appropriate fields into the category may reduce memory space. This is true for text data of low cardinality (with many “repeats” in its data range); data with a large percentage of totally unique values will not benefit from such conversion.
Conversion can also boost computational efficiency as string transformations will be performed on the categories attribute rather than on every individual element of the dataframe. In other words, the change is done once per unique category, and the results are mapped back to the values.
Let’s explore the dataset.
print(df.info()) >>> RangeIndex: 126169 entries, 0 to 126168 Data columns (total 7 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 sex 126169 non-null object 1 lifestage 126169 non-null object 2 scientificName 126169 non-null object 3 eventDate 126169 non-null object 4 geoprivacy 126169 non-null object 5 recordedBy 126169 non-null int64 6 stateProvince 126169 non-null object dtypes: int64(1), object(6) memory usage: 6.7+ MB
Dataset has 6 columns with object dtype.
len(df.sex)/df.sex.nunique() >>> 126169.0
In the sex column, the ratio of values for every unique value (category) is 126169. In other words, the column has only 1 unique value. This is so, as, in frog species, typically male frogs call, while females stay silent. Such a high ratio makes this column a great candidate for converting into a categorical type.
To convert column to category df.sex = df.sex.astype('category')
To measure memory used df.col_name.memory_usage(). It returns memory usage in bytes.
print(f'before: {df.sex.memory_usage(index=False, deep=True)}')
df.sex = df.sex.astype('category')
print(f'after: {df.sex.memory_usage(index=False, deep=True)}')
>>>
before: 7696309
after: 126310 # 98% improvement
While converting to categorical dtype promises memory savings, the result may not always be such dramatic. As has been mentioned above, the effectiveness depends on the ratio of values for every unique category. However, in our example, converting ‘sex’ values into category alone slashed memory usage by 98% (from 7.34 Mb to 0.12 Mb)!
And if we decide to rename ‘sex’ values for something more descriptive, this will be done only once for each category, not on every element of 126169 elements of the dataframe.
The other candidates for converting to categorical are ‘stateProvince’, ‘scientificName’, ‘lifestage’, and ‘geoprivacy’ columns with the ratio of 15771.1, 685.7, 126169.0 and 63084.5 values for every unique category, respectively.
We can streamline converting objects to a category by abstracting the process into function. To avoid changing columns with dates, we pass such column names as a parameter to ignore them for now. Also, we abstract coefficient ‘c’ to help filter out columns worth converting to ‘category’ dtype.
def optimize_object(df: pd.DataFrame, datetime_cols: list, c=0.5) -> pd.DataFrame:
for col in df.select_dtypes(include=['object']):
if col not in datetime_cols:
if (df[col].nunique()/len(df[col])) < c:
df[col] = df[col].astype('category')
else:
pass
return df
Numerical dtypes
Pandas integer types include int8, int16, int32, int64, uint8, uint16, uint32, uint64> with int64 being default type for scalar values.
But int64 is able to hold values in range from negative Nine quintillion two hundred twenty-three quadrillion three hundred seventy-two trillion thirty-six billion eight hundred fifty-four million seven hundred seventy-five thousand eight hundred eight to the same amount on positive side minus one. In case of the frogs dataframe ‘recordedBy’ column only has values in range from 19 to 36155. So casting to lower integer type can save space. As values are all positive, using unsigned integer is right choice.
print(f'before: {df.recordedBy.memory_usage(index=False, deep=True)}')
df.recordedBy = df.recordedBy.astype('uint16')
print(f'after: {df.recordedBy.memory_usage(index=False, deep=True)}')
>>>
before: 1009352
after: 252338 # 75% improvement
Max and min values for integer types
Type Values Bytes int8 ... -128 to 127 ... 1 uint8 ... 0 to 255 ... 1 int16 ... -32,768 to 32,767 ... 2 uint16 ... 0 to 65,535 ... 2 int32 ... -2,147,483,648 to 2,147,483,647 ... 4 uint32 ... 0 to 4,294,967,295 ... 4 int64 ... -9.223372e+18 to 9.223372e+18 ... 8 uint64 ... 0 to 1.844674e+19 ... 8
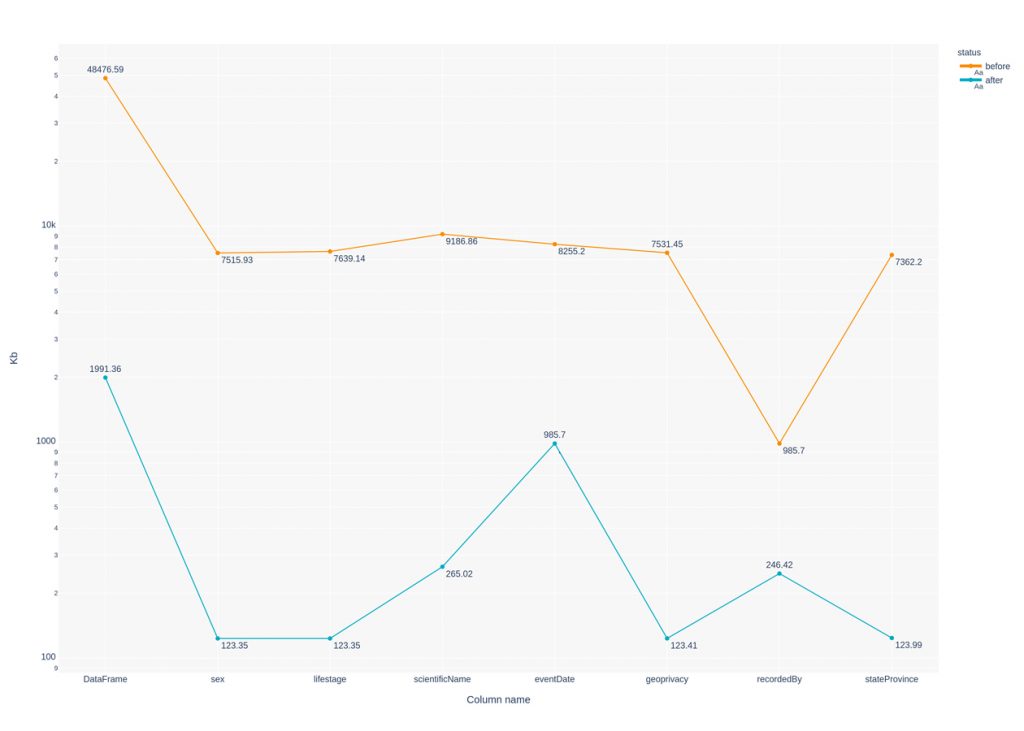
The following graph shows in logarithmic progression memory usage for each of the columns and entire DataFrame before and after datatype optimization. eventDate column has been converted to datetime: df.eventDate = pd.to_datetime(df.eventDate).
After transformation dataframe is looking like that:
RangeIndex: 126169 entries, 0 to 126168
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 sex 126169 non-null category
1 lifestage 126169 non-null category
2 scientificName 126169 non-null category
3 eventDate 126169 non-null datetime64[ns]
4 geoprivacy 126169 non-null category
5 recordedBy 126169 non-null uint16
6 stateProvince 126169 non-null category
dtypes: category(5), datetime64[ns](1), uint16(1)
memory usage: 1.9 MB
Note on int and Int types in Pandas
To put it simply, ‘Int’ types are for representing integer data with possibly missing values.
Pandas, if unspecified, uses NaN to represent missing data. But because NaN is float type, this forces an array of integers with any missing values to become an array of floats. For cases where casting an integer to float can be problematic, pandas offers Int type. Int is not an integer but an integer array of pandas extension types. When converting values to ‘Int’, all NA-like values will be replaced with pandas.NA rather than numpy.nan.
Resources:
Python Pandas: Tricks & Features You May Not Know
What is the difference between native int type and the numpy.int types?
Pandas DataFrame: Performance Optimization
How to optimize your Pandas code